Working to exploit insertional mutagenesis for cancer gene discovery since 1996.
Research projects in the Largaespada lab are aimed at identifying mutations and other changes that drive the development of cancer, which must be determined for developing molecularly targeted therapeutics.
Sleeping Beauty
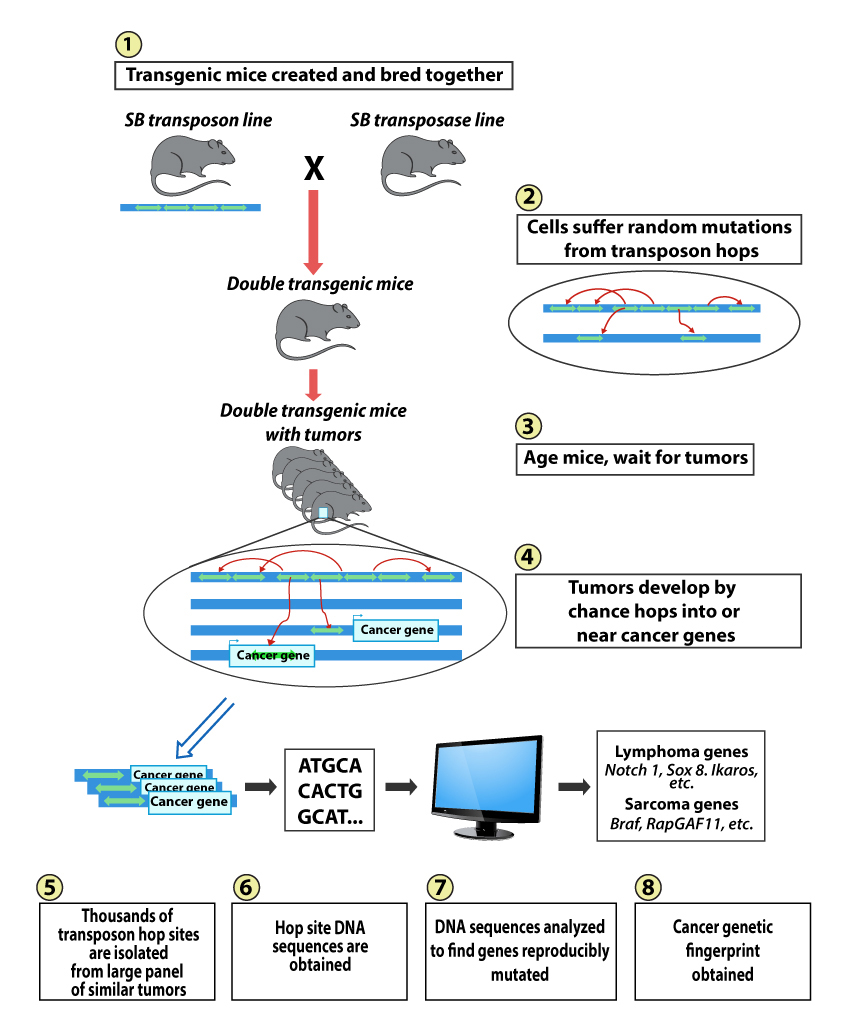
The Largaespada lab pioneered the use of a vertebrate-active transposon system, called Sleeping Beauty (SB), for insertional mutagenesis in mouse somatic cells. SB is being used as a tool for forward genetic screens for cancer genes involved in sarcoma, hepatocellular carcinoma, mammary, gastro-intestinal tract and NF1 syndrome-associated nervous system cancers.
Novel Mouse Models
Also, novel mouse models are being used for preclinical evaluation of new drugs and drug combinations for cancer treatment. A large focus is on drugs to treat tumors associated with Neurofibromatosis Type 1 (NF1) syndrome.
Molecular Biology
The lab is also working to utilize and improve the latest methods in genome engineering, like CRISPR/Cas9, to study cancer gene function and uncover cancer vulnerabilities.
Research Vision
Genes and pathways important for cancer relevant phenotypes are being sought using functional genomics approaches. In particular, the lab has invested heavily in the use of Sleeping Beauty (SB) transposon mutagenesis in mice to create various types of cancer and to reveal novel cancer genes at recurrently mutated loci. These models and phenotypically and genetically similar to human cancers, allow meaningful genotype-phenotype correlations to be drawn, and reflect human cancer genetic heterogeneity within and between different tumors. We have applied deep RNA sequencing to transposon induced tumors and can easily find fusion transcripts between the transposon and endogenous genes. These analyses have revealed new cancer genes missed by only studying transposon insertions at the level of genomic DNA. These data also allow genotype-phenotype correlations to be drawn finding downstream gene expression changes for specific gene alterations. Follow-up studies on these genes and pathways involve genetic engineering of normal human cells using targeted nucleases such as CRISPR/Cas9. Genes from our forward cancer genetic screens using SB mice are being knocked out in normal immortalized human cells and cancer cell lines. These studies are revealing what cancer phenotypes these genes control. We are using engineered cell line models, and genetically engineered mouse models, to test the effects of pharmacological interventions, and for genetic synthetic lethal studies. Improved methods for transposon mutagenesis, genome editing and genome manipulation in general are being pursued.