The human genome is poised to reshape clinical medicine
Time passages are marked in our DNA. Since the completion of the Human Genome Project (HGP) was announced 20 years ago, each of us has acquired, on average, about 60 single-letter DNA mutations, a low rate compared to other species. Most are not important. Genetic changes that are important to our well-being, whether we acquired them since 2003, since we were born, or inherited them from our parents, are less and less able to stay concealed. That is HGP’s enduring legacy and a springboard for genomic medicine.
What is genomic medicine? The National Human Genome Research Institute (NHGRI) defines it as an emerging medical discipline that involves using genomic information about people as part of their clinical care. Although genomic medicine is already making its mark in the fields of cancer, rare and infectious diseases, pharmacogenomics, and prenatal testing, LMP professor Bharat Thyagarajan and his colleagues are determined to expedite its broader use in the clinic.
Thyagarajan is director of the department’s Advanced Research and Diagnostic Laboratory  (ARDL). The timing of ARDL’s Genomics Core initiative, which Thyagarajan leads, is auspicious for at least three reasons:
(ARDL). The timing of ARDL’s Genomics Core initiative, which Thyagarajan leads, is auspicious for at least three reasons:
- The power of long-read DNA sequencing to identify structural changes in our genome and measure our exposure to environmental risks, which is ARDL’s current focus
- Our ability to penetrate and analyze single cells and their regulatory networks in exquisite molecular detail, a major advance over bulk cell averaging for identifying drug targets, and
- Generative artificial intelligence or generative AI being brought to bear on the information pathways and networks of the human body, aiding disease diagnosis, therapy, and prevention.
Does all this mean we’ve reached a tipping for genomic medicine, that soon it will enter patient care and healthcare more broadly in a very big way?
2003-2023: Amazing speed, lag times and tipping points
The International Human Genome Sequencing Consortium announced the completion of the Human Genome Project in April 2003, more than two years ahead of schedule, under budget, and 50 years to the month after James Watson and Francis Crick published “A Structure for Deoxyribose Nucleic Acid” in the journal Nature.
Thyagarajan remembers that he was a University student working on his doctorate in cancer epidemiology when a draft of the human genome sequence was published in Nature in 2001. The journal’s cover featured the iconic DNA double helix composed of a crowd of human faces including those of Watson, Crick, and Gregor Mendel, the father of modern genetics.  He recalls that his class discussed the paper and what it might mean, the potential benefits for research and clinical medicine. What has surprised him most in the ensuing years?
He recalls that his class discussed the paper and what it might mean, the potential benefits for research and clinical medicine. What has surprised him most in the ensuing years?
“The speed at which things have changed,” Thyagarajan said in an interview. “When I was finishing up my PhD, it was just around the time we were in the initial phase of doing genome-wide association studies.” According to NHGRI, a genome-wide association study (abbreviated GWAS) is a research approach used to identify genomic variants, typically single-nucleotide variations or SNPs, that are statistically associated with a risk for a disease or a particular trait. “I thought, okay, this is going to be a research activity,” he said. “I wasn't thinking about clinical applications at that point.”
With the introduction of next-generation sequencing (NGS) in 2009, Thyagarajan thought, “Oh, cool! You knew this was going to replace all the research applications for genome-wide association studies, which it did. But if you had asked me then whether we would have seen this widespread adoption of next-generation sequencing and clinical work, I would have said ‘absolutely not.’ There were too many errors in sequencing. Plus, it was so difficult to analyze the data.”
Due to those limitations, Thyagarajan was doubtful genomics would play a prominent role in the clinic anytime soon. But the accelerating speed, greater accuracy, and declining costs of whole genome sequencing (WGS) in which all or nearly all of an individual’s 3.3 billion DNA base pairs are identified have altered his outlook.
The core of the Core
ARDL’s Genomics Core began to take shape two years ago with a proposal to leverage its national leadership as the long-standing central biochemistry laboratory for NIH-sponsored multicenter longitudinal cohort studies. It set out to expand its testing capabilities to include genomic analysis of incoming specimens and to work both with partners in industry and within the M Health Fairview System to develop novel tests. To fulfill the Core’s mission, ARDL acquired short- and long-read DNA sequencing and digital PCR (polymerase chain reaction) instruments with their reagents, created an in-house bioinformatics capability, and hired new personnel.
Thyagarajan said ARDL’s Genomics Core initiative will address “two broad areas” that make full use of its scientific and technical capabilities:
- Adding value to existing and future population-based cohort studies, and
- Establishing ARDL as a genomics reference laboratory through industry partnerships to develop genomics-based tests for the marketplace.
On the genome sequencing front, laboratories focus on speed, accuracy, and cost. Speed and accuracy have steadily improved. A human genome costs ARDL $200 to sequence (analysis and annotation add to the cost) compared to about $3 billion to sequence the first human genome, with the cost continuing to drop.
“But just saying we will do standard whole genome sequencing is no longer enough,” Thyagarajan said, noting that through NIH development programs 200,000 human genomes are already available. “You have to think about what additional value we can bring to genomics and through genomics to patient care.”
That’s where long-read sequencing, what Thyagarajan calls third-generation sequencing, comes in. Not only does the technology enable investigators to identify structural changes in the genome, it can also enable them to identify epigenetic changes to the DNA bases – changes that reflect an individual’s environmental and occupational exposure. Epigenetics “allows you to get a broader picture of environmental exposures, using genomics as a backbone,” he said.
Epigenetic changes are key contributors to the exposome, defined as the measure of all the 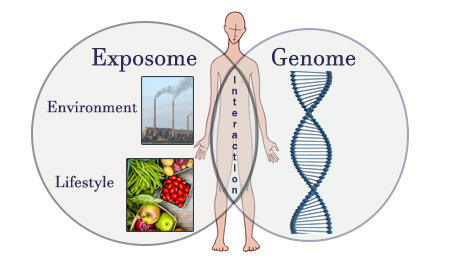 exposures of an individual in a lifetime – exposures from where one lives, where one works, the air one breathes, one’s diet and lifestyle, etc. -- and how those exposures relate to health. Genomics plus other “omics” such as proteomics -- the large-scale study of proteins -- and microbiomics -- the study of the community of organisms living inside each of us -- can reveal informative exposome biomarkers.
exposures of an individual in a lifetime – exposures from where one lives, where one works, the air one breathes, one’s diet and lifestyle, etc. -- and how those exposures relate to health. Genomics plus other “omics” such as proteomics -- the large-scale study of proteins -- and microbiomics -- the study of the community of organisms living inside each of us -- can reveal informative exposome biomarkers.
Given ARDL’s four decades as a central reference laboratory for multi-institutional clinical studies and trials for diabetes, cardiovascular disease, kidney disease, and other conditions, its vast specimen biobank, and its recent leadership role in COVID-19 diagnostic and antibody testing, few if any laboratories are better prepared to do what ARDL’s Genomics Core is setting out to do. In addition to the samples already on hand, ARDL is collecting new samples for sequencing including from large social science as well as epidemiological cohort studies.
ARDL’s genome sequencing strategy of cohort populations dovetails with its intention of becoming a leading genomics reference laboratory through industry partnerships to develop genomics-based tests. “We are making investments not just in long-read sequencing technology but also in technologies that will enable us to promote partnerships with industry and help them get their tests into the market,” Thyagarajan said, adding that he envisions ARDL as becoming a reference lab for facilitating genomics-based diagnostic test development and U. S. Food and Drug Administration (FDA) approvals.
ARDL has ongoing interactions with companies developing diagnostic tests for lung cancer, minimal residual disease (MRD) detection in blood cancers, asthma, and other conditions. It is also partnering within the M Health Fairview System, Thyagarajan said. ARDL has been working with both MDL (Molecular Diagnostic Laboratory) and IDDL (Infectious Disease Diagnostic Laboratory) to develop long-read sequencing tests for molecular diagnostics (MDL) and microbiome diagnostics (IDDL). “We’ve made substantial progress in both those areas,” he said. “I anticipate, hopefully yet this year, that we will implement some of these as clinical tests in the M Health Fairview System.”
New tools for a new era
Perhaps the most formidable tools in genomic medicine in the years ahead never touch bodily fluids or mark the contours of a cell, a tissue, an organ, or a body. They are literally the disembodied and cell-free tools of machine learning (ML) and artificial intelligence (AI) with their data training sets and predictive algorithms. Yet the future of genomic medicine, with its innate biological complexity, will depend on how safely and effectively ML and AI can be applied in the diagnosis, treatment, and prevention of disease.
Building ML and AI models that can predict gene expression from DNA sequences has been a long-standing research goal, especially for the development of RNA-based therapies. The big picture, however, sees multimodal AI combining and analyzing data from medical records,  genomics and other omics including of single cells, medical imaging and other pattern recognition technologies, and wearable sensors that use telemetry to signal underlying physiological conditions plus ambient air quality and the environment—the exposome and social determinants of health. Large language models (LLMs) that drive generative AI programs such as ChatGPT (Microsoft) and Bard (Google) are making their way into medical research and practice, including at ARDL.
genomics and other omics including of single cells, medical imaging and other pattern recognition technologies, and wearable sensors that use telemetry to signal underlying physiological conditions plus ambient air quality and the environment—the exposome and social determinants of health. Large language models (LLMs) that drive generative AI programs such as ChatGPT (Microsoft) and Bard (Google) are making their way into medical research and practice, including at ARDL.
“I actually encourage my students to use generative AI while writing papers, even for drafting comments on patients,” Thyagarajan said, stressing the enhanced productivity such tools offer. “We must figure out how to use generative AI to improve our productivity. I think that is the key thing.” He reflected that if in the 1990s some investigators decided they were not going to use Microsoft Word, “You're just not going to be competent. You wouldn't have been competitive for very long. And I think the same thing applies here.”
Thyagarajan uses Google Bard as an assist tool – a hybrid approach -- in doing genomic interpretation and differential diagnosis, the process of differentiating between two or more conditions that share similar signs or symptoms. “I put in patient symptoms, ask what the differential diagnosis is, what is recommended, and it gives pretty good results,” he said.
What does Thyagarajan hope to see in 20 years, at the 40th anniversary of the HGP’s completion?
“I’m hoping that in the next 20 years, all these investments and advances in diagnostics like pan-cancer screening will be getting translated into improvements in patient care and patient treatments,” he said. It may take that much time, but he believes the time will come.
In the research arena, the ability of investigators to generate information far outstrips their ability to convert that information into knowledge, he said. “There is so much omics data already available in the public domain. And yet, we have not been able to take all that information and synthesize it to advance our understanding of biology. We are in the infancy of figuring out how we can use multi-omics data to take the next step.”
That step invariably will combine ideas and ingenuity, which ARDL has demonstrated over the decades, with AI, which borrows from the unparalleled connectivity and signal processing of the human brain, a creative force like none other.
---------
The image at the top of this story is from the cover of the February 15, 2001 edition of Nature entitled “the human genome.”



