Sun pinpoints a key divergence in the relentless logic of cancer’s spread
In his Pulitzer Prize-winning book The Emperor of All Maladies: A Biography of Cancer, Columbia University physician Siddhartha Mukherjee describes cancer as a clonal disease because all known cancers arise from one ancestral cell. “But cancer is not simply a clonal disease; it is a clonally evolving disease,” he writes. “If growth occurred without evolution, cancer cells would not be imbued with their potent capacity to invade, survive, and metastasize.”
Cancer, Mukherjee contends, exploits the “fundamental logic of evolution” unlike any other disease because every generation of cancer cells creates a small number of cells that is genetically different from its parents. A population of mutated cells with greater growth potential may then leave the tumor microenvironment and travel to distant tissues and organs through the blood and lymph systems.
The fundamental logic of cancer has come under scrutiny by a new generation of scientists equipped with new tools and methods. Some call themselves evolutionary oncologists.

One of them is LMP assistant professor Ruping Sun, a faculty member in the Masonic Cancer Center’s Genetic Mechanisms Program, which was set up to define the genetic changes that initiate cancer and drive its progression. Sun is an expert in cancer genomics and the computational modeling of cancer – in innovating advanced bioinformatic solutions for measuring cancer evolution from tumor genomic sequencing data.
Recently, Sun has focused his expertise on discerning the natural history of metastatic cancer, the evolutionary “branching” of the tumor from its seedbed of primary cancer cells. Such branching is a standard feature of phylogenetic trees, family tree-like diagrams that show the evolutionary relationships among species or, in this case, cells.
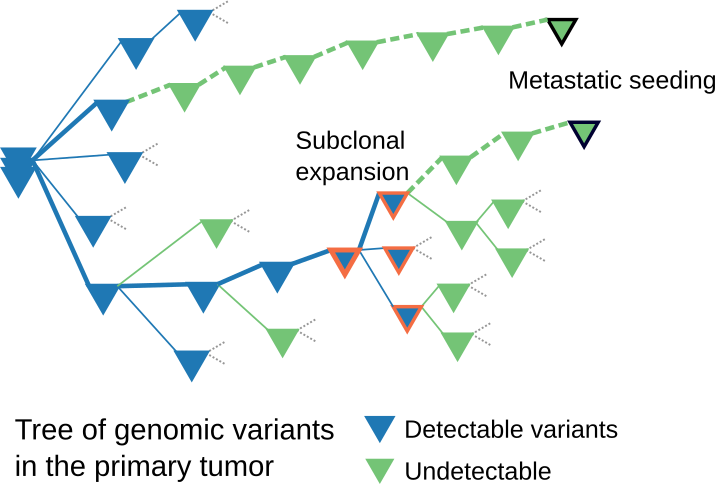
Reconstruction of the phylogenetic tree of cancer metastasis through modeling can help reveal the key factors involved in cancer progression and guide clinicians in diagnosing and treating patients.
In March, Sun and LMP research scientist Athanasios Nikolakopoulos published “Elements and evolutionary determinants of genomic divergence between paired primary and metastatic tumors” in PLoS Computational Biology. In light of the greater focus on analyzing the phylogenetic trees between paired metastatic and primary tumor samples, they asked the question “Can metastatic-primary (M-P) genomic divergence measured from next-generation sequencing reveal the natural history of metastatic dissemination?”

The Cancer Genome Atlas (TCGA), established in 2006 by the National Cancer Institute and the National Human Genome Research Institute, has characterized more than 20,000 primary cancers at the molecular level for 33 different tumor types. That effort has led to improvements in cancer diagnosis, treatment, and prevention.
With the better understanding of genomic abnormalities in primary tumors, the cancer genomics community is moving toward pinpointing the roots of cancer progression by characterizing the genomic differences between longitudinal tumor samples – samples analyzed over a period of time. The exact meaning of the measured genomic divergence on cancer evolutionary trees, however, has been elusive. Metastatic seeding times of similar genomic tree patterns have yielded different interpretations.
Sun and Nikolakopoulos wanted to know how and when a primary cancer cell, typically localized in a tissue microenvironment, evolves into a metastatic clone, a cell population with the capacity to spread. Using computational and mathematical modeling of cancer progression and virtual genomic profiles, they set out to illuminate the conditions under which metastatic-primary (M-P) divergence patterns reveal fundamental evolutionary parameters, such as the timing of metastasis.
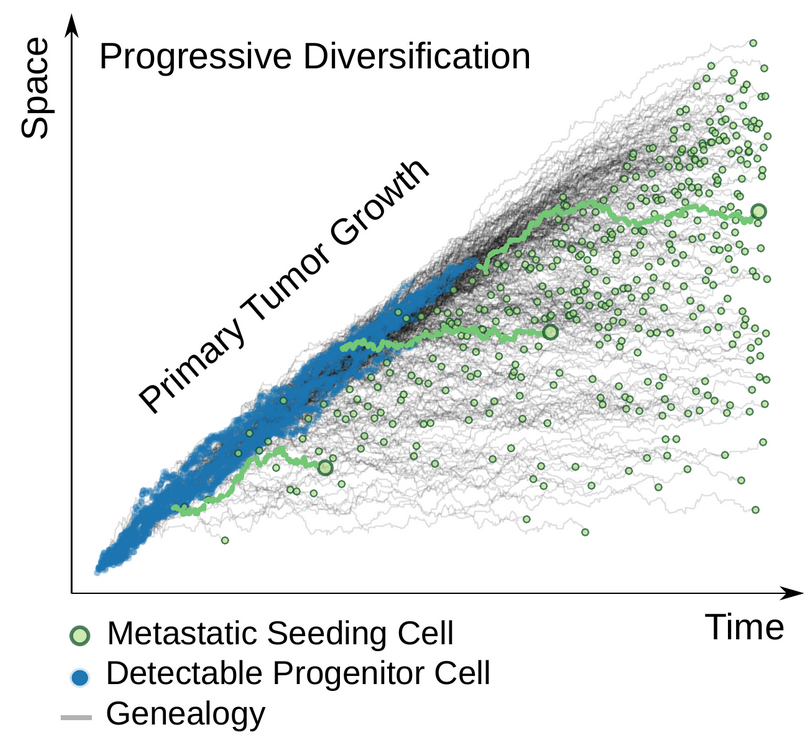
They succeeded. By analytically reducing the measurable M-P divergence to its intrinsic components, they uncovered quantitative rules about how tumor growth modes, metastatic seeding times, driver subclone decisions in metastatic seeding, and sequencing detectability of variants all combine to regulate the M-P divergence observed in next-generation DNA sequencing (NGS) experiments. As they report in their paper, that enabled them to create a framework that “can serve as a foundation for future data-driven efforts aimed at understanding more complex metastatic dynamics.”
Sun said in an interview that he and Nikolakopoulos did not focus on a particular type of cancer. “We built a theoretical framework that is applicable to cancer in general including solid tumors and blood cancers.” Their M-P divergence pattern framework also applies to general between-tumor analysis, such as the comparison between pre- and post-treatment samples, he said.
The “fundamental logic of cancer” can take years or decades to play out. It often results in a heterogeneous tumor made up of cell populations with distinct characteristics. The process is launched by a “founder cell” in a primary tumor, a cell possessing driver mutations that contribute to one or more cell populations with different features and potential to spread. An evolving clonal descendant population of the founder tumor cell typically acquires additional (epi)genetic alterations that equip it with the ability to leave the primary site and initiate an “invasion-metastasis cascade,” usually resulting in diverse and genetically heterogeneous tumor cell populations. Sun said his research illuminates the correlation between the primary tumor expansion and the relative timing of the metastatic seeding of these cell populations, based on next-generation DNA sequencing (NGS) data. The cancer evolutionary tree is cast into sharp relief.

Because single tumor cells under analysis may differ depending on the precise location of biopsied tissue, in a parallel study Sun and Nikolakopoulos constructed a single cell-based tumor simulation that allowed them to represent the spatial heterogeneity of the tumor. In the spatial simulation they found the same M-P divergence pattern, corroborating their mathematical predictions, which assume a well-mixed cancer cell population, Sun said.
Metastatic cancer often features bulk genomic changes such as extra or missing chromosomes, called aneuploidy, and copy number variation, in which sections of individual chromosomes are duplicated or repeated. These genomic features complicate the kind of DNA sequence-based model building Sun and Nikolakopoulos have undertaken. Though they haven’t formally incorporated these types of changes into their theoretical framework, they have introduced “an interface from which we can bridge this information into our model,” Sun said. “Right now in the cancer evolution field people often neglect to account for copy number changes in measuring evolutionary parameters.”
One of Sun’s near-term goals is to use his theoretical M-P divergence framework to build what he calls a user-friendly “lookup table” that can account for the extent of whole genomic changes arising from pathway divergences. He calls the table an “inspirational research tool” that will improve steadily over time with access to more data and a better quantitative understanding of the dynamics of cancer progression.
The Cancer Genome Atlas provides a lookup table to assist users in navigating the Genomic Data Commons Data Portal of primary cancers that arise from a single ancestral cancer cell and confine their growth to the primary anatomical site.

But it is the relentless logic of tumor evolution taking malignant cells to a breakaway divergent path and metastatic disease that earns cancer the title of Siddhartha Mukherjee’s book: The Emperor of All Maladies.