Spatialomics
Requesting Training & Services
If you are interested in Spatialomic services please submit a UMN Spatialomics Project Interest Form to get started. A member of the UMN Spatialomics team will be in touch to discuss your project.
A Collaborative Frontier
University of Minnesota Spatialomic services are partnership between the University Imaging Centers (UIC) and the University of Minnesota Genomics Centers (UMGC).Together, we work in concert to provide new avenues for researchers to increase their understanding of cells in their morphological context. The relationship between cells and their relative locations within a tissue sample can be critical to understanding disease pathology. Spatialomics allows scientists to measure all the gene activity in a tissue sample and map where the activity is occurring. Already this technology is leading to new discoveries that will prove instrumental in helping scientists gain a better understanding of biological processes and disease.
Spatial Technologies
MIBIscope
MIBI
The MIBIscope System is a revolutionary imaging platform, enabling comprehensive phenotypic profiling and spatial analysis of the tissue microenvironment. The MIBIscope allows researchers to visualize over 40 markers simultaneously with higher sensitivity, resolution, and throughput than existing methods.
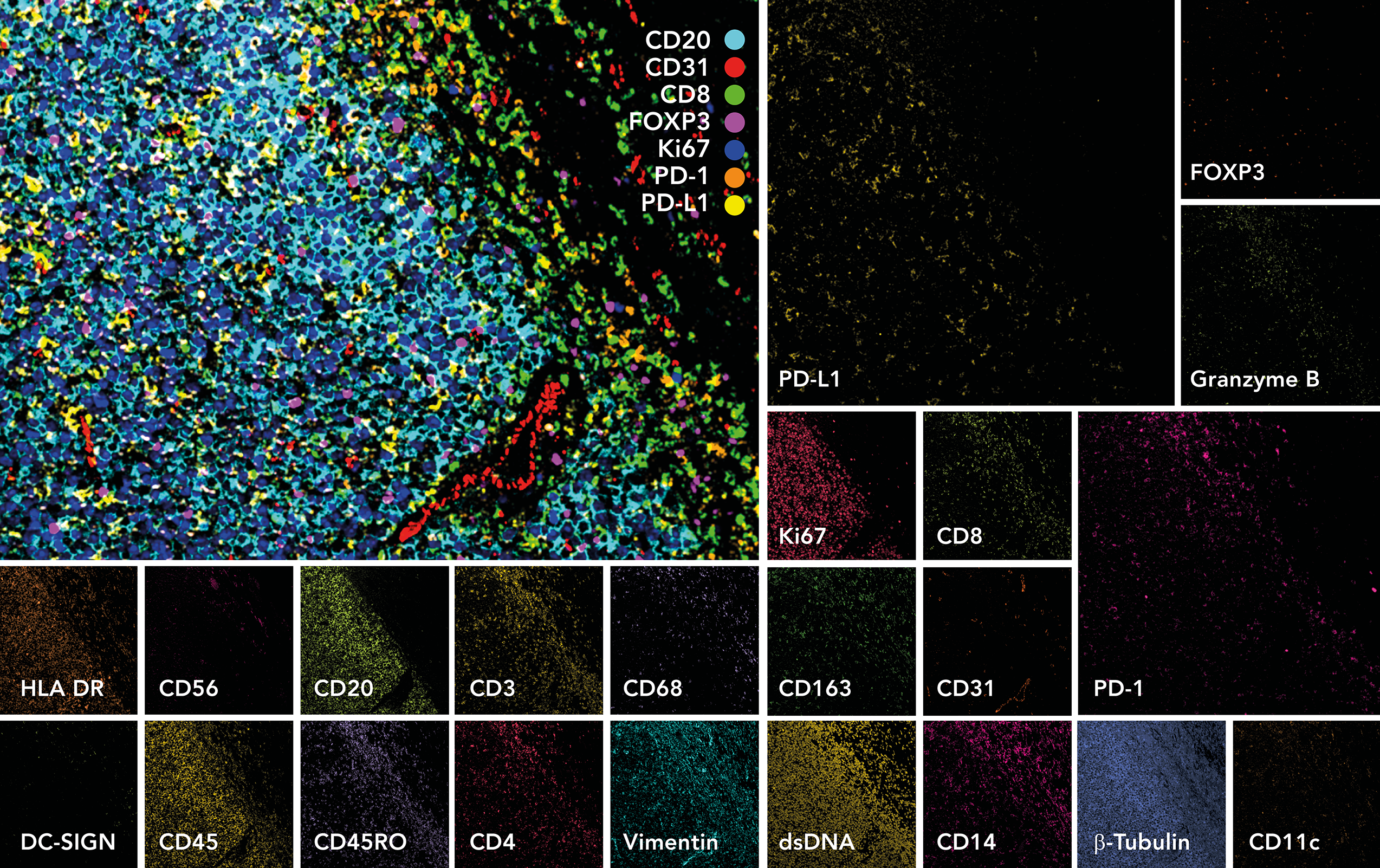
MIBI Technology is based on secondary ion mass spectrometry or SIMS. With SIMS, a primary ion beam is rastered across the surface of a sample, liberating reporter ions that are then simultaneously recorded on a pixel-by-pixel basis by Time-of-Flight detection. An ion beam, unlike a laser, enables resolution to be tuned over a broad range—in the case of the MIBIScope, from a few hundred nanometers to 1 micron. Once liberated, the reporter ions, or “secondary ions,” travel uninterrupted at supersonic speed from the sample to the detector, leading to fast acquisition and extraordinary sensitivity.
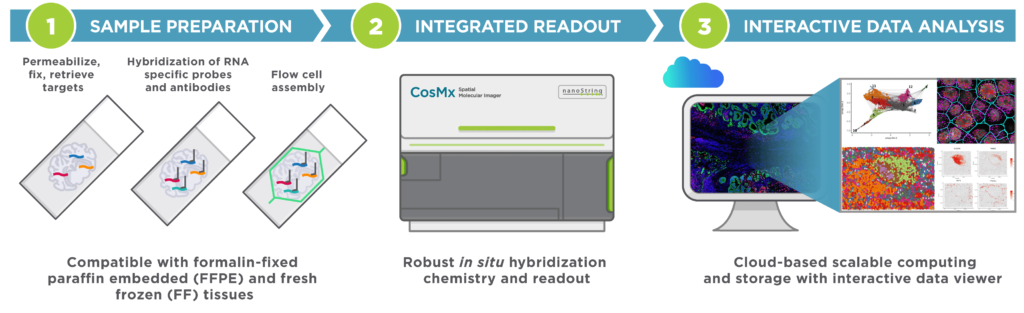
CosMx Spatial Molecular Imager (SMI)
CosMx Spatial Molecular Imager (SMI)

CosMx SMI (Spatial Molecular Imager) is a high-plex in situ analysis platform providing spatial multiomics with formalin-fixed paraffin-embedded (FFPE) and fresh frozen (FF) tissue samples at cellular and subcellular resolution. CosMx SMI enables rapid quantification and visualization of up to 1,000 RNA and 64 validated protein analytes. It is the flexible, spatial single-cell imaging platform that will drive deeper insights for cell atlasing, tissue phenotyping, cell-cell interactions, cellular processes, and biomarker discovery.
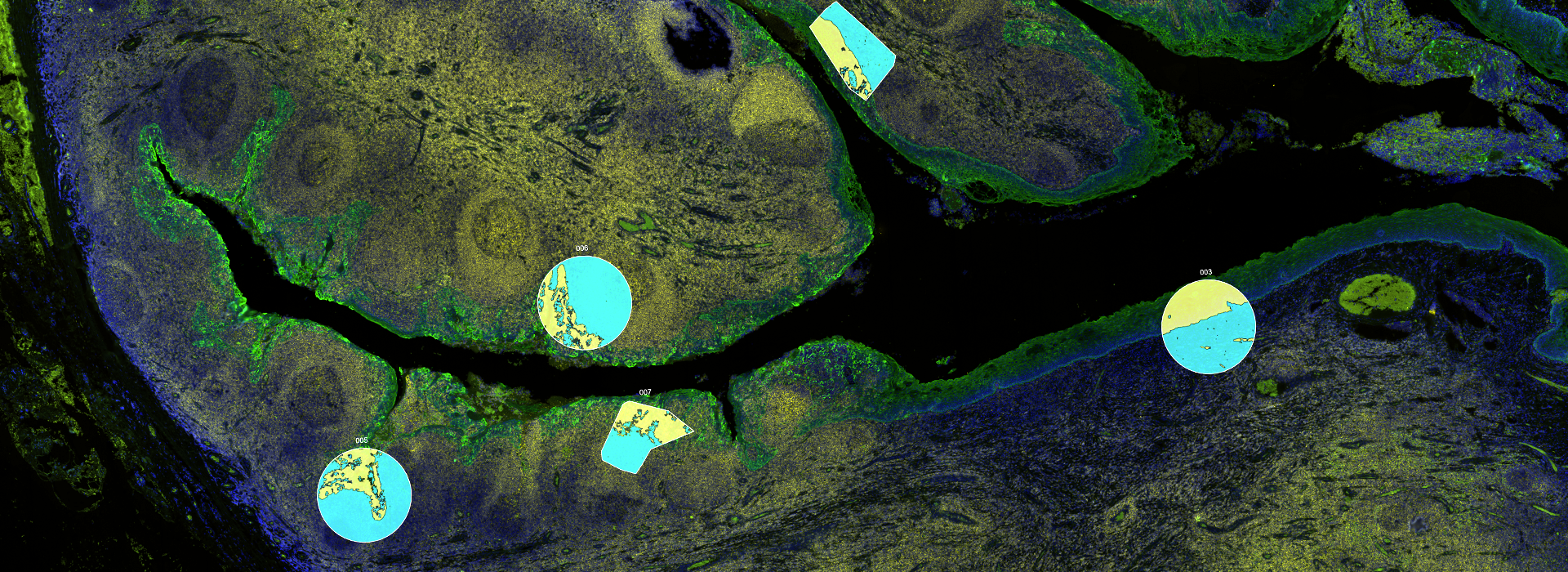
GeoMx Digital Spatial Profiling (DSP)
GeoMx Digital Spatial Profiler (DSP)

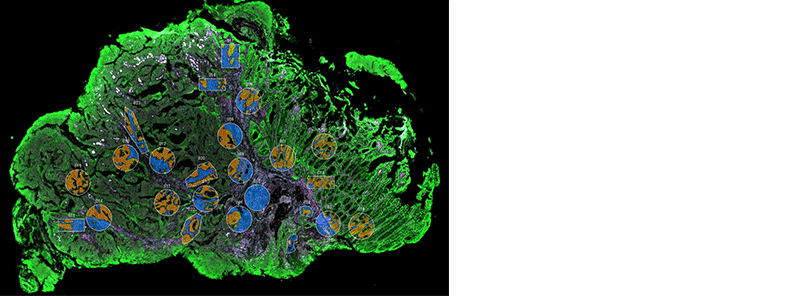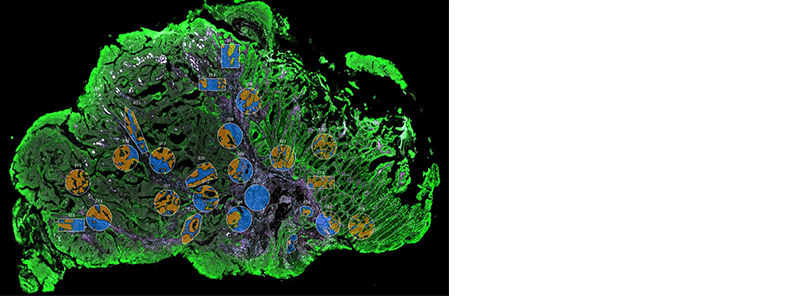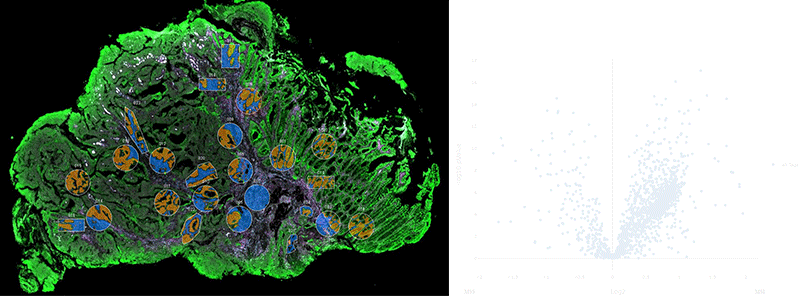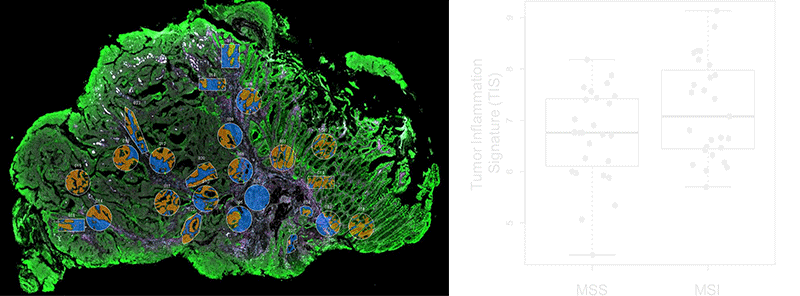
NanoString’s GeoMx Digital Spatial Profiler (DSP) combines standard immunofluorescence techniques with digital optical barcoding technology to perform highly multiplexed, spatially resolved profiling experiments. In a single reaction, the DSP technology performs whole slide imaging with up to four fluorescent stains to capture tissue morphology and select regions of interest for high plex profiling. The ability to perform tissue morphology guided profiling experiments increases the likelihood of capturing rare events often missed by typical grind and bind assays. The DSP chemistry enables spatially resolved high plex profiling of RNA and protein targets on just two serial sample sections. The ability to perform multianalyte analysis enables deep characterization of the sample.
The DSP combines the best of spatial and molecular profiling technologies by generating a whole tissue image at single-cell resolution and digital profiling data for 10’s-1,000’s of RNA or Protein analytes for up to 4 tissue slides per day including FFPE. This unique combination of high-plex, high-throughput spatial profiling enables researchers to rapidly and quantitatively assess the biological implications of the heterogeneity within tissue samples.