Tissue Clearing
Requesting Training and Services
If you are interested in gaining access to UIC resources or requesting services please follow the steps on our training and service request page.
Overview
The University Imaging Centers integrates state-of-the-art tissue clearing methods and technologies to help investigators prepare, image, and analyze biologically intact tissues and organs in three dimensions.
Tissue clearing brings a new dimension to histology services, where historically, tissue sectioning not only provides limited information of biological structures in its native form, but also proved to be a laborious and computationally intensive challenge in reconstruction from 2D to 3D. An assortment of tissue clearing methods has been developed in multiple laboratories within the last decade to make various tissues optically transparent by reducing light scattering intrinsic in tissues. This enables optimized whole-mount imaging on our specialized imaging systems, in particular, the Caliber ID RS-G4 Ribbon Scanning Confocal and the 3i Cleared Tissue Light Sheet (CTLS) microscope.
The UIC employs multiple tissue clearing approaches while primarily focusing on two: the PEGASOS organic-based and X-CLARITY hydrogel-based methods. PEGASOS and X-CLARITY are compatible with antibody and fluorescence staining, respectively, which allows for a comprehensive analysis of preserved internal structures.
SmartBatch+
The UIC’s SmartBatch+ system combines hydrogel-based tissue clearing and active immunolabeling. Tissue processing for the SmartBatch+ includes an initial step of additional tissue fixation using SHIELD reagents to protect endogenous fluorescence and tissue structure. Following SHIELD fixation and hydrogel matrix formation, the SmartBatch+ uses a rotating electric field to pull SDS micelles through samples, effectively removing the lipids while maintaining tissue dimensions and architecture. Once samples are delipidated, the SmartBatch+ labeling process uses stochastic electrotransport to produce thorough labeling of intact tissue volumes.
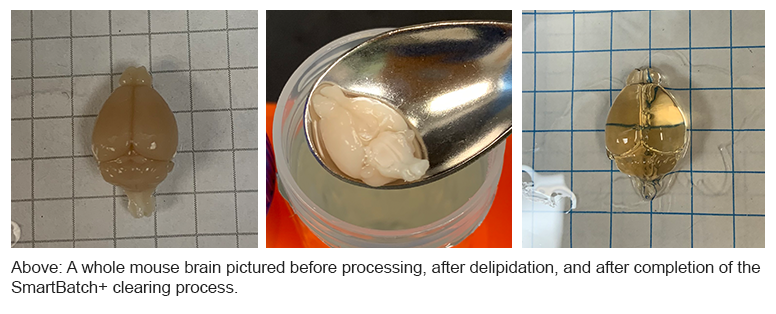
This method allows for the use of almost tenfold less antibody than traditional passive staining of whole tissues. Contact the UIC to determine whether your chosen antibodies are validated for use with the SmartBatch+ system.
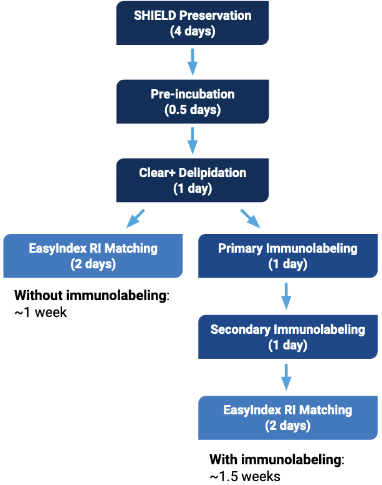
For samples such as whole mouse brains or similarly sized tissues, the entirety of the clearing and immunolabeling process can take as little as 1.5 weeks with the SmartBatch+. Further, this device is optimized for batch processing with up to 12 mouse brains that can be cleared in a single run. Samples cleared with the SmartBatch+ system are then mounted in a refractive index-matching solution optimized for imaging on the Caliber ID RS-G4 or the 3i CTLS at the UIC.
LifeCanvas Validated SmartBatch+ Antibodies
X-CLARITY
With the X-CLARITY method, the tissues are immobilized within a hydrogel and lipids are removed by electrophoresis in the presence of a detergent. This approach stabilizes the tissue while preserving proteins and nucleic acids throughout with very small, isotropic changes in tissue dimensions. While this method works exceptionally well with endogenous fluorescent proteins, the density of the hydrogel matrix can make antibody labeling of larger samples challenging.

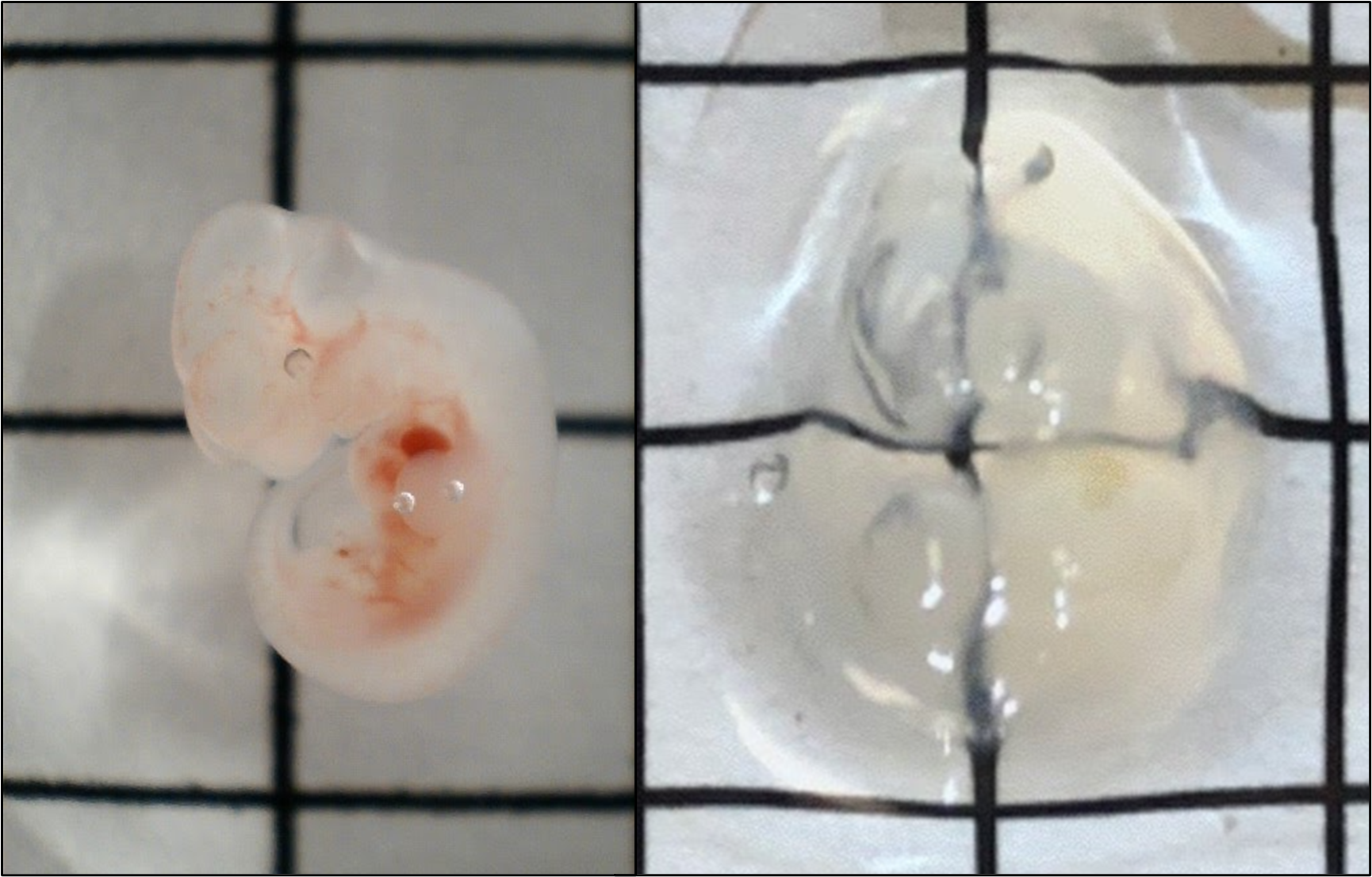
Above: Images of an intact mouse brain and spinal cord before and after X-CLARITY clearing. Grid squares = 1 x 1 cm.
PEGASOS
Compared to other organic-based methods, PEGASOS is capable of antibody labeling while preserving endogenous fluorescence. The PEGASOS method employs organic solvents to solubilize lipids and render tissues transparent once in refractive index matching reagents. PEGASOS is our method of choice for projects in which post-fixation dye, staining and/or immunolabeling is required. This method preserves endogenous fluorescent protein signals for extended times as well and optimally creates a uniform refractive index required for light-sheet imaging.
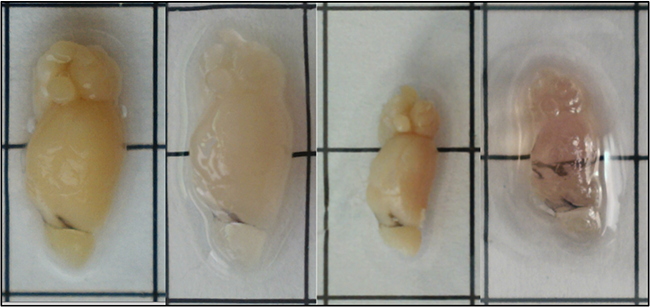
Above: Images of a mouse hemibrain (7-mm thickness) processed using the PEGASOS clearing method. The steps from left to right include 4% PFA fixation, delipidation, dehydration, refractive index matching. Grid square = 1 x 1 cm.
Above: From the laboratory of Dr. Amy Yi-Mei Yang, the University Imaging Centers at the University of Minnesota presents cleared tissue light-sheet imaging of td-Tomato-labeled parvalbumin interneurons in a mouse cerebrum and hippocampus segment. The tissue was both cleared by PEGASOS and imaged on the 3i Cleared Tissue Light-Sheet microscope by the University Imaging Centers
Fixation and sample preparation for submission
Perfuse mice transcardially with 1X PBS, then 4% PFA, followed by tissue extraction and immersion in 4% PFA in 1X PBS for 24 h at 4˚C. Tissue(s) should then be washed with 1X PBS and stored in PBS at 4˚C. The PFA should be freshly made from EM grade or better stock.
References
Clearing for Deep Tissue Imaging
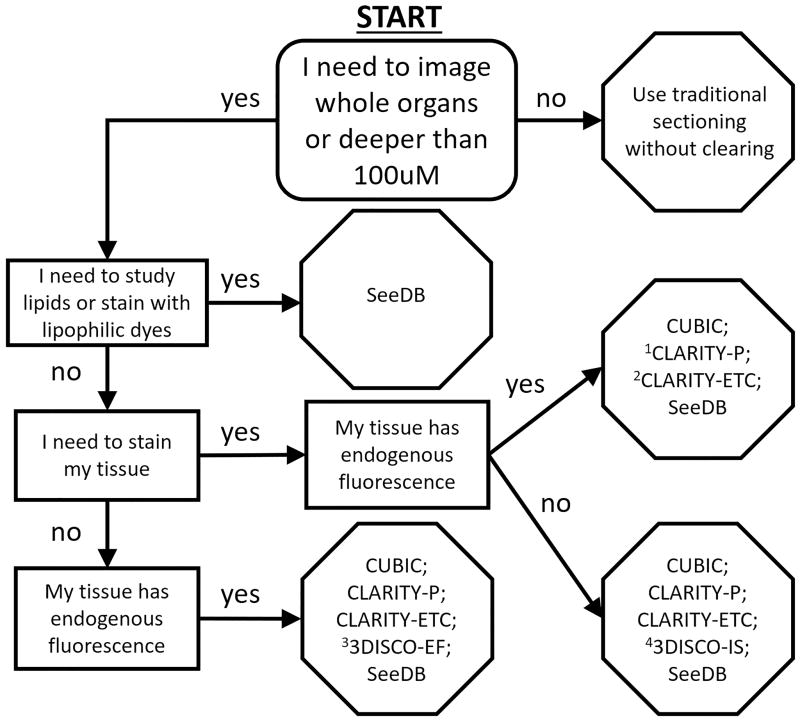
Clarifying Tissue Clearing
Tissue Clearing
Tutorial: practical considerations for tissue clearing and imaging
Authors: Kurt R. Weiss, Fabian F Voigt, Douglas P. Shepherd & Jan Huisken
Summary
These two methods provide the opportunity to reveal the relationship between the structure and function of molecules of interest in an intact 3D organization for imaging in plant and animal tissues. While there are many other tissue clearing methods, and we are happy to help you pursue those, we have found these two methods work well with most biological and our imaging systems. We are happy to work with you to determine which system and which probes are right for your studies.
Project Pipeline
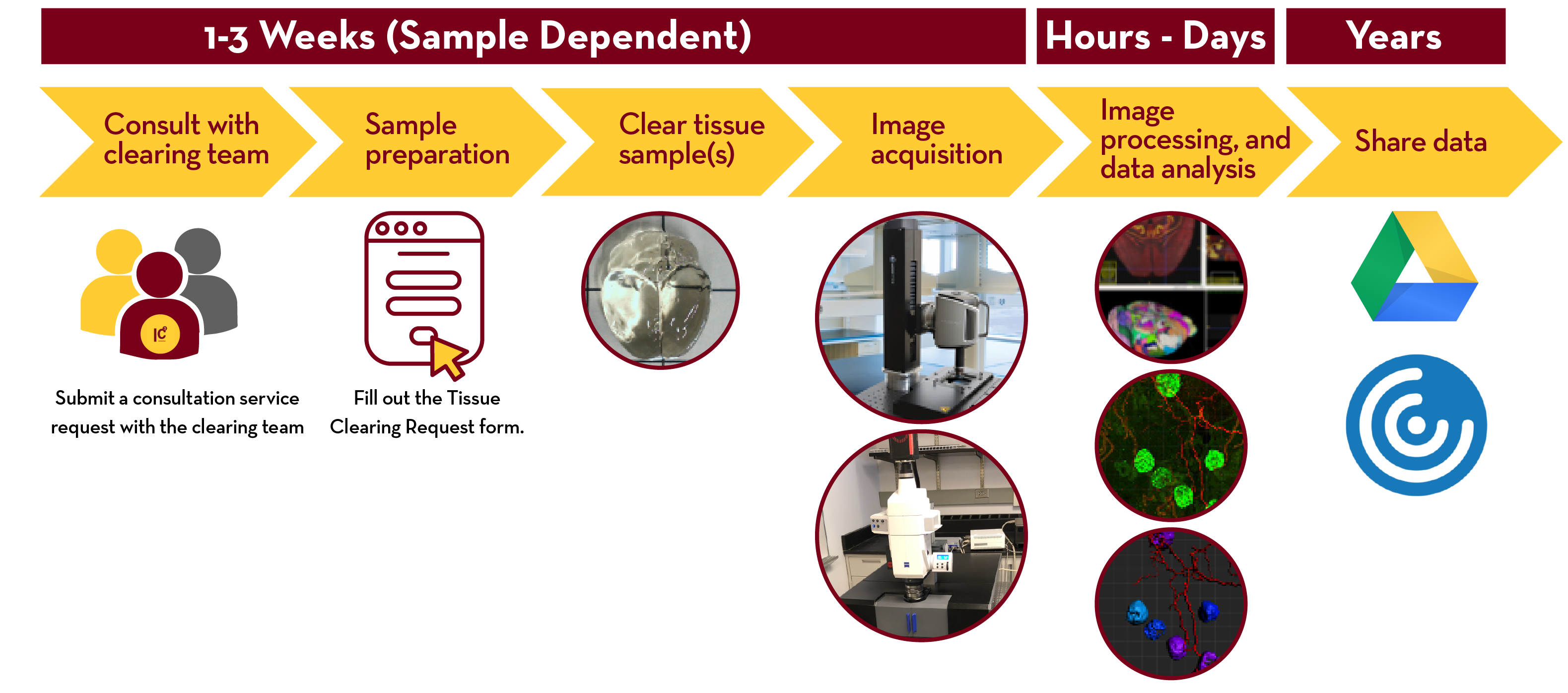
To initiate or discuss a project, please submit a service request. Next, clearing team members will set a time to consult with the client regarding sample preparation, the fluorescent labeling, required validation steps and goals for imaging and data analysis. The client then submits a clearing request via UIC Tissue Clearing Request Form Submission. A sample submission date will then be agreed upon and the client will notify the team when sample preparation and fixation occurs in advance. The fixed tissue sample(s) can be dropped off at any of the UIC’s facilities. The clearing team confirms receipt of the samples and logs the samples into our database and begins the processing. Depending on the tissue size, tissue type, labeling requirements, and the clearing method, the whole process may take 1-4 weeks. Once completed, clearing staff coordinates a time to preview each sample on either the 3i Cleared Tissue Light-Sheet (CTLS) microscope for PEGASOS-cleared samples or the Caliber RSG4 ribbon-scanning confocal microscope for PEGASOS-cleared samples or X-CLARITY-cleared samples to confirm labeling and determine the resolution and ROI for final imaging and then begins imaging that sample. (we would be excited to not have to do this step remotely!). Once the imaging is completed (one or more days/sample), the acquired images are registered and converted to an Imaris format for ease of viewing these large datasets and shared with the client via a secure RDS server. Clearing staff is to notify the client for them to help the client remotely explore the data using a VM with Imaris viewer. The client can then confirm the image and schedule the discussion of image analysis and visualization with Thomas, James or UIC staff.
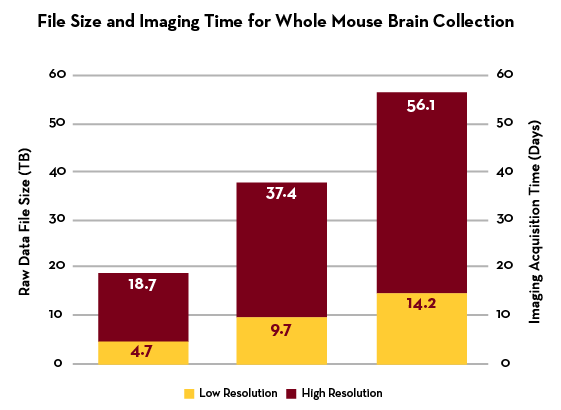
This graph illustrates the relationship of how file size and image acquisition time scales for an average intact mouse brain-sized samples post clearing (14 x 18 x 5 mm) taken on the Caliber ID RSG4 Ribbon Scanning Confocal with the SLWD 20X/1.00 NA Glyc objective. The number of channels and the resolution have a significant impact on collection times, data size and subsequent processing and analysis efforts. The numbers on the bars represent the estimated file sizes for raw data in terabytes (TB) on the left axis corresponding to imaging acquisition time measured in days on the right axis. The average data size for an intact mouse brain ranges from 2-3 TB. Conversely, PEGASOS cleared samples imaged on the CTLS ranges from 600 GB - 1 TB. *Note there is shrinkage in tissues processed by the PEGASOS method.
Imaging Systems
CTLS
- Sample dimensions range: 10mm x 10mm x 10mm
- Lasers: 488 nm, 561 nm, 515 nm, 640 nm
- Optimal with PEGASOS and other non-hydrogel methods
RS-G4
- Sample dimension range 30mm x 30mm x 10mm
- Lasers: 405 nm (not recommended), 488 nm, 561 nm, 647 nm, and 785 (Reflectance Laser)
- Optimal with X-CLARITY and PEGASOS methods
Resonant and Galvo Confocals (A1RHD MP, A1Rsi HD W/SIM, A1R-FLIM, C2+ Spectral)
-
Not as fast as the RSG4 and CTLS modalities, but can go to higher resolution and spectral collection as needed