Structural Circuits Core
The Structural Circuits Core offers state-of-the-art anatomical mapping of neural circuits through automated use of brain clearing technology paired with meso- and micro-scale imaging of the central nervous system
The SCC offers cutting-edge anatomical mapping of neural circuits involved in addiction. Integrated with the University Imaging Center and utilizing the UMN Informatics Institute, the SCC also provides infrastructure for automated use of brain clearing technology paired with meso- and micro-scale imaging of the CNS, coupled with computational approaches to mine information from the resulting large sets of data. This Core will provides investigators with a set of powerful tools to establish the anatomical connectivity of discrete elements of neural circuits (e.g. neurons that express a specific molecular marker or activate an immediate-early gene during a particular behavioral epoch). These elements can be genetically tagged with a fluorophore, but antibody labeling in cleared, whole-brain tissue is also an option (or a combination). An important feature of the SCC is the capability to generate powerful quantitative datasets by registering sets of individually imaged brains from a given treatment condition.
cleared tissue light-sheet imaging of td-Tomato-labeled interneurons in a mouse cerebrum and hippocampus
From the laboratory of Dr. Amy Yi-Mei Yang, the University Imaging Centers at the University of Minnesota presents cleared tissue light-sheet imaging of td-Tomato-labeled parvalbumin interneurons in a mouse cerebrum and hippocampus segment. The tissue was both cleared by PEGASOS and imaged on the 3i Cleared Tissue Light-Sheet microscope by the University Imaging Centers
Tissue Clearing and Imaging Pipeline
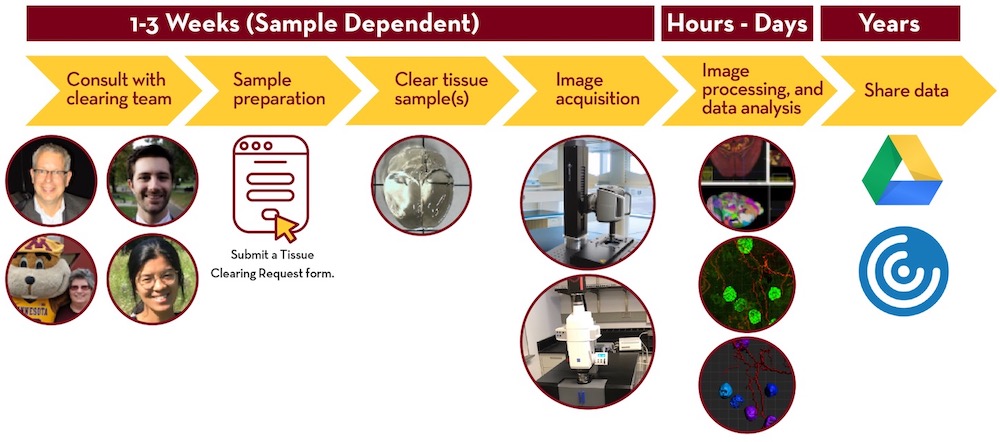


Potentially include additional text on mouse brain and spinal cord
Structural Circuits Core Leads and Team

Paul Mermelstein, PhD Dr. Mermelstein is a Professor in the Department of Neuroscience in the University of Minnesota Medical School. Dr. Mermelstein co-leads the SCC with Dr. Heilbronner and with assistance from the director of the University of Minnesota's Imaging Center, Dr. Mark Sanders, and the Associate Director of the UMN Informatics Institute, Dr. Thomas Pengo.

Sarah Heilbronner, PhD Dr. Heilbronner is an Associate Professor at Baylor College of Medicine. Dr. Heilbronner co-leads the SCC with Dr. Mermelstein and with assistance from the director of the University of Minnesota's Imaging Center, Dr. Mark Sanders, and the Associate Director of the UMN Informatics Institute, Dr. Thomas Pengo.
Mark Sanders, PhD Dr. Sanders is the program manager for the UMN Imaging Center (UIC). He provides leadership in the formulation, development and implementation of the University-wide shared multi-scale microscopy facility.
Thomas Pengo, PhD Dr. Pengo is the Director of Applications and Services in the UMN Informatics Institute (UMII). His role is both to provide direction for UMII and to provide image analysis services for any researcher at UMN. His background in computer engineering and he has over a decade of experience in image analysis makes him ideally suited for guiding the development and coordination of the computational platform.
James Zhang James Zhang has a strong background is in Computer Science and Engineering, with expertise in artificial intelligence, robotics and web development. His role in the SCC is to provide development and operational support to the image analysis workflows in collaboration with Dr. Pengo.
For additional information on tissue clearing or imaging systems see the UMN Imaging Center website.
Contact Information:
For general inquries about instrumentation, services, or projects please send a message at uic-staff@umn.edu. Once received, a member of the UIC team will followup to discuss your inquiry.
Additional information on Data Analysis:
Imaging Informatics: Imaging Informatics services now include image analysis across many different imaging modalities. From fluorescence microscopy, to small animal imaging, neuroimaging and material science. Dr. Thomas Pengo provides analysis support for biological images (any instrument at the University Imaging Centers). Services include 2D image analysis (morphometry, particle analysis, densiometry, etc.) and 3D image analysis (volumetric sample measurement) reconstruction and image editing. Analysis is performed with FIJI, QuPath, MATLAB, Nikon Elements, Imaris, Amira, MATLAB and Python software.Analysis tools include open-source software and customized analysis pipelines.
Contact: Thomas Pengo, Ph.d., tpengo@umn.edu. Office: 1-220 CCRB